Samples of STING
Millennium application:
| Fig_1. |
|
Fig_2. |
|
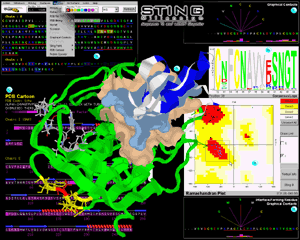
In figure 1 we show
an artistic collage of snapshots of different modules of STING
Millennium session that analyzed alpha-chymotrypsin complex with
turkey ovomucoid third domain ( 1cho.pdb); We opted to present images
and features accompanied by insets (A through F, in Fig
1 and A through D, in Fig 2) and general
description of menu and features that a user can find behind each inset
presentation: |
Central image is structure of 1cho.pdb already carrying information from
actions taken during analysis. Green ribbon is enzyme:
chymotrypsin chain and white one is inhibitor: ovomucoid third domain. We have
prepared this image by executing following STING Millennium
commands/features: differential coloring of present structural chains,
ribbon presentation [see QuickLearn Action menu
example]; calculation of the interface forming residues (IFR) [see Interface
on example] ; calculation and display of the IFR surfaces (gold surface belongs
to IFR's of the E chain and cyan surface belongs to IFR's of I chain) [see QuickLearn
Build Surface and Interface example]; selection of 8 residues from the E chain
sequence (white background in sequence window) and subsequent presentation in
wireframe + spacefill (ws) mode in 3D with
CPK coloring of atoms [see mouse marking of
contiguous sequence region example]; selection of a single amino acid and
coloring it in red (red background in sequence window); calculating all neighbors
(from a selected residue) included within a sphere of radius of 5 Å from/to Last
Heavy Atom (LHA) in a side chain of each amino acid (color coded yellow both in
structure and sequence window)[see QuickLearn
Search Pattern and Search Neighbors example]; color coding in blue of I chain
ribbon for all amino acid residues selected by Ramachandran plot (see inset D
on Fig. 1)[see QuickLearn Ramachandran example].
Sequence and control window of STING Millennium :
It displays linear protein and/or DNA sequence, color coded according to hydrophobicity
and charged groups [see Sequence Color Coding].
The sequence window also shows the numbering of the residues in the sequence [see
Residue Numbers], gaps in the PDB sequence
[see Sequence Gaps in PDB file], a chain identifier
[see Sequence Chains] and ranges for secondary
structure elements [see Secondary Structure
Regions]. Each residue in the sequence window is "clickable",
resulting in a presentation of its position in the structure window (there are
9 colors to be chosen from [Residue Color option
on control panel] and 7 different display options
[menu bar left from Refresh button on control panel] : Wireframe, WS =Wireframe
+ Spacefill, CPK, Ribbon, Backbone, Strands and Cartoon) . Blue
and Red lines below the sequence are also "clickable" resulting in graphical ribbon
presentation of the specified sequence region (red lines
indicate helical region and blue
lines indicate extended sheets region). STING Millennium
Status Frame [right from STING's icon (bee) on control panel] shows the
residue/nucleotide number and chain identification
any time a user slides the mouse over a residues/nucleotides on the sequence window
or any time a user clicks the mouse over a certain atom in the structure window.
In the sequence window, background of a residue
single letter code changes color accordingly to the chosen: Residue Color. Selecting
residues by clicking or sliding the mouse above the sequence will cause background
color to change. White background residues (VPGSWPWQ) marked in the Sequence Window
are displayed in a CPK color presentation in the Structure Window. Parity and
easy localization in sequence and space coordinates
is fully implemented in this way. STING Millennium
Action menu displays STING script commands, the purpose of which is to perform
a simple structure analysis: it color codes all charged
residues or differentially colors all chains,
selects and displays ligand, displays ligand and water molecules in vicinity of
the ligand, selects and displays ligand
pockets and finally, offers "copy to clipboard" command that facilitates transfer
of STING Millennium image to adequate program for image manipulation (like PhotoShop
etc.). STING Millennium Windows menu offers more
complex commands: Interface chain, surface,
search pattern, neighbors and send script.
This last one makes possible direct issuing of Rasmol/Chime commands to Structure
Window and is aimed to more experienced users.
PDB Cartoon is a image of the amino acid
sequence along with the secondary structure elements rendered as cartoons. In
this case user has chosen to see temperature factor (provided by PDB file), which
is color coded as background of single letter amino acid code. Interesting off-spin
of this presentation is usable information of how good is a PDB definition of
secondary structure elements: residues R-230 and V-231 of chain E, are both in
the helix and in the extended sheet (also seen in sequence window annotation).
PDB Cartoon can show background amino acid single letter code to represent physico
chemical characteristics (rather than temperature factor as in shown inset C on
fig. 1) [see also QuickLearn Pdb Cartoon
example].
Ramachandran plot is displaying the main-chain
dihedral angles (Phy and Psi). In this inset, we have
chosen to see only "I" chain angles. In addition, we have selected (blue color
dots) one of allowed regions and then map them on 3D structure window (also blue
colored ribbon) by using "Sting it!" button. Most of the chosen residues are forming
helical structure. Blue colored amino acids in the Structure Window are also identified
by blue background (chain I) in the Sequence Window [see also QuickLearn
Ramachandran Plot example].
Contacts : This inset shows fan-like virtual
contact lines coming out from residue Y-11 (this numbering counts for 3 missing
amino acid residues at the very N terminal) of chain I and points to an amino
acid that makes a particular contact (identified by the color of the line connecting
it). Information about the distance, atom partners and the type of the contact
is provided on context and if a particular residue is selected with the mouse,
then a zoomed image is displayed in the Structure Window representing the contacting
environment in detail. The histogram along sequence of the chain I, aids in rapid
localization for critical residues, having larger than average number of contacts
and also, buy color differentiation, having more energy-valuable contacts (like
electrostatic interaction) [see also QuickLearn
Graphical Contacts example].
Interface Forming Residues Contacts: this
inset demonstrates the same sequence of the chain I, but now the user can see
IFR as red underlined regions. Crucial difference from the inset E, is that here,
contacts are counted only between residues belonging to the different chains.
One can easily spot that chosen residue L-18 of chain I, makes number of different
contacts with the E chain, while it makes no contact with residues of its own
I chain (see inset E on Fig. 1)[see also QuickLearn
IFR Contacts example] .
Consensus Sequence: One of the STING Millennium
features is a capacity to extract valuable data from HSSP database and then represent
them in a variety of usable ways, such as this at inset G. Here the user can quickly
grasp how diversified is a sequence for the family of protease inhibitors that
have high sequence homology. In this inset, the larger the letter the larger is
conservation at that particular position in sequence. However, real valuable information
comes from the displayed variation of amino acids that can occupy this position.
For example, position 40 is occupied by glutamic acid (in inset B on Fig. 1, one
can follow this sequence region as helical VVE, which is in the last row, left
side). That very same position can also be occupied by aspartic acid, but most
interestingly, it also shows that some inhibitors have "accepted" a mutation that
replaced negatively charged residue with positively charged one: both arginine
and lysine. Exact percentages for amino acid type that occupies specific position
are available interactively from this module.
|
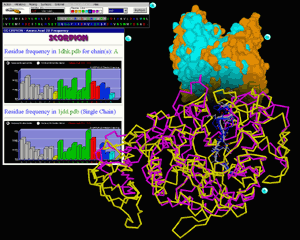
On figure 2 we present exceptionally useful features
of STING Millennium: display of multiple structure alignment in 3D and
also sequence alignment (presented in sequence window). The sequence alignment
follows in this case structural alignment. Structure can then be color
coded with respect to calculated sequence entropy which consequently offers
to the user a visual tool for easy identification of spots exposed to
the differential selective pressure.
|
In this inset we show structural superposition (done by using PRISM
program) of two amylase enzymes: porcine pancreatic alpha-amylase (1dhk.pdb
in yellow backbone) and maltotetraose-forming
exo-amylase (1jdd.pdb in magenta backbone).
Although two enzymes have extensive differences if total fold is compared, there
are striking similarities inside the active site. We have marked differentially
two sequence regions with very good 3D overlap and which are forming respective
active sites. White backbone belongs to 1dhk.pdb and blue one belongs to 1jdd.pdb.
[See also QuickLearn Modes example]
Sequence window here has a crucial difference from the one presented in Fig
1.; Namely, STING Millennium here presents two
sequences in such an order so that they follow structural alignment. Top sequence
is the one of 1dhk.pdb and the bottom one is of the 1jdd.pdb. Differentially
marked background colors are following same logistics as previously described
for Fig.1. A user can easily spot conservation along the marked sequences. [See
also QuickLearn Modes example]
Scorpion is a program that provided graphs
for this inset. We present here residue frequency for the whole molecule (1dhk.pdb
on top and 1jdd.pdb on bottom graph). This information can be transformed in
cumulative frequencies for hydrophobic, polar and charged groups (one click
away from demonstrated graph). Differences in frequencies are visually much
easier to analyze than otherwise. [See also QuickLearn
Scorpion example]
Surfaces of two superimposed molecules are presented on this inset. Such presentation
visually aids in analysis of the active site (V shaped opening on the top of
the figure). Golden color surface belongs to 1dhk.pdb and cyan colored one is
of the 1jdd.pdb. [See also QuickLearn Build
Surface example]
NOTE: before
closing this session, check-out how you can add even more details to this rich
structural description produced exclusively by SMS by using appropriate data
parsing {active SMS links}!