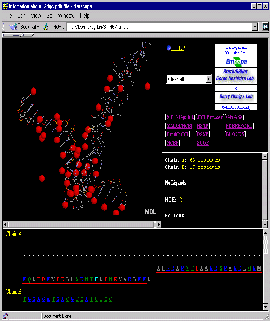
Press:{Shift left mouse button}: enlarge image a little!
Observe: in the Sequence Frame that sequence of the A
chain starts with residue number 229, causing STING to show long stretch
of "------" as indication of missing residues!
Press: [Charged residues] button on STING control panel
Observe: all charged residues are colored on the protein chain,
clearly indicating positively charged moiety engaged in DNA binding.
Curiously, there is also negatively charged Glutamic acid in this region:
E, chain A,#237.
Slide: mouse over sequence residues at Sequence Frame!
Residue numbers will appear on the STING's Status Frame!
Note that STING handles well negative residue numbers
for DNA chain
Press:[HOH off]!
Press:[Color by chain]!
Press:[Interface on]!
Press:[HOH+Interface]!
Observe:water molecules in-between protein and DNA surface,
clearly involved in H-bond network formed among two molecules!
Click:(Sequence Frame) on B chain, nucleotide G,#1.
Observe:this is a central nucleotide of the DNA bound stretch!
Final Screen: You will get screen as shown below:
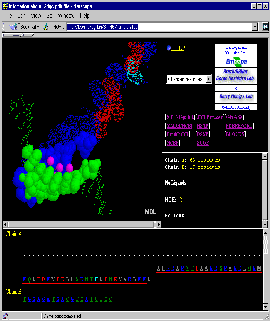 And Beyond: Use [Interface: 1st half] and
[Interface: 2nd half] to get exclusive view on first half and then
second half of the complementary Interface Forming Residues (IFR)!
Continuing, use [Interface: 1st half] to get
again exclusive look at the first half of the interface...
Such iterative procedure allows users to get better glimpse at IFR.
Very useful STING command will be [Charged residues]: user can see
how charge is distributed and how complemented at Interface halves!
And Beyond: Use [Interface: 1st half] and
[Interface: 2nd half] to get exclusive view on first half and then
second half of the complementary Interface Forming Residues (IFR)!
Continuing, use [Interface: 1st half] to get
again exclusive look at the first half of the interface...
Such iterative procedure allows users to get better glimpse at IFR.
Very useful STING command will be [Charged residues]: user can see
how charge is distributed and how complemented at Interface halves!
STING:
Ligand Pocket and Coordinating Residues
In this example, we will look at the Ligand Pocket within Protein Fold
Step by step instructions
First of all, open the pdb file 1ytc, by typing in
this 4 letter code and then pressing "Enter" key on the keyboard,
at entry STING page
(simply click on STING It icon, and you will be in familiar area).
You will get screen as shown below:
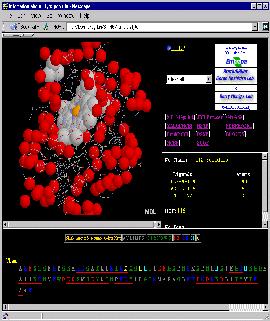 Press:{Shift left mouse button}: enlarge image a little!
Turn it to get most pleasing view on the molecule!
Observe: in the Sequence Frame: there are 3
Histidine residues! Click on each of them to see 3D position with respect to
HEME. Clearly, HIS #18 is the one coordinated to Fe of the HEME ligand!
Press: [Charged residues] button on STING control panel
Observe: all charged residues are colored on the protein chain,
clearly indicating Histidine residue (cyan color) engaged in HEME
(LIGAND) binding.
Slide: mouse over sequence residues at LF! Residue numbers
will appear on the browser's lower border! Observe HIS sequence numbers!
Again, HIS #18 is the one coordinated to Fe of the HEME ligand!
Press:{Shift left mouse button}: enlarge image a little!
Turn it to get most pleasing view on the molecule!
Observe: in the Sequence Frame: there are 3
Histidine residues! Click on each of them to see 3D position with respect to
HEME. Clearly, HIS #18 is the one coordinated to Fe of the HEME ligand!
Press: [Charged residues] button on STING control panel
Observe: all charged residues are colored on the protein chain,
clearly indicating Histidine residue (cyan color) engaged in HEME
(LIGAND) binding.
Slide: mouse over sequence residues at LF! Residue numbers
will appear on the browser's lower border! Observe HIS sequence numbers!
Again, HIS #18 is the one coordinated to Fe of the HEME ligand!
Note that STING handles well negative residue numbers
for protein chain!
Press:[ribbon] button on STING control panel!
Press:[ligand off]
Press:[ligand on]
Observe: Histidins placed appropriately around Ligand for coordination.
Press:[Ligand Pocket]
Press:[ligand off]
Press:[ligand on]
Observe: only atoms in contact with LIGAND are presented in
CPK graphics format.
|
Click: with mouse on each HIS (cyan) residue on the Graphics
Frame.
Observe: within STING's Status Frame residue number!
|
Final Screen: You will get screen as shown below:
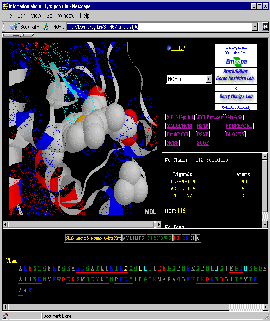
STING: Enzyme/Inhibitor Interface
and Structural Waters
In this example, we will look at the Enzyme Inhibitor Interface
Step by step instructions
First of all, open the pdb file 1slu, by typing in
this 4 letter code and then pressing "Enter" key on the keyboard,
at entry STING page (simply
click on the STING It icon, and you will be in familiar
area).
You will get screen as shown below:
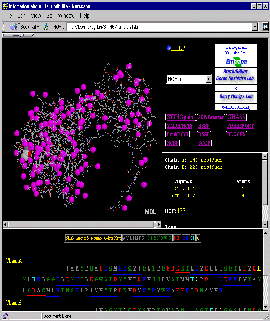 Press:{Shift left mouse button}: enlarge image a little!
Observe: in the Sequence Frame that both chains
do not start with residue number 1, causing
STING to show a stretch of "------" as indication of missing residues!
Also present are gaps in the sequence, easily identified in the
Sequence Frame.
Slide: mouse over sequence residues at Sequence Frame!
Residue numbers will appear on the STING's Status Frame!
Press:[HOH off]!
Press:[Ligand Pocket]
Press:[HOH + Ligand]
Adjust: for the best view!
Press:[ligand off]
Press:[ligand on]
Observe: Ligand pocket and residues creating it,
as well as structural water molecules.!
Press:[Color by chain]
Press:[Interface on]!
Press:[HOH+Interface]!
Observe:water molecules in-between Enzyme and Inhibitor surface,
some of which are involved
in H-bond network formed among two molecules!
Click:Sequence Frame Arginin on B chain, residue #96.
Observe: position of this residue w/r/t interface!
Click:(UR) frame, pointer BLOCKS
Type_in: CHYMOTRYPSIN in the separate BLOCKS browser search area!
Click: on BLOCKS browser Edit menu option and then type in
TRY1 at FIND option!
Observe: following sequence profile: ( 43) GGSLINDQWVVSAAHC
Click: on boxes bellow GGSLINDQWVVTAAHC sequence, starting at
position B#43.
Press:{Shift left mouse button}: enlarge image a little!
Observe: in the Sequence Frame that both chains
do not start with residue number 1, causing
STING to show a stretch of "------" as indication of missing residues!
Also present are gaps in the sequence, easily identified in the
Sequence Frame.
Slide: mouse over sequence residues at Sequence Frame!
Residue numbers will appear on the STING's Status Frame!
Press:[HOH off]!
Press:[Ligand Pocket]
Press:[HOH + Ligand]
Adjust: for the best view!
Press:[ligand off]
Press:[ligand on]
Observe: Ligand pocket and residues creating it,
as well as structural water molecules.!
Press:[Color by chain]
Press:[Interface on]!
Press:[HOH+Interface]!
Observe:water molecules in-between Enzyme and Inhibitor surface,
some of which are involved
in H-bond network formed among two molecules!
Click:Sequence Frame Arginin on B chain, residue #96.
Observe: position of this residue w/r/t interface!
Click:(UR) frame, pointer BLOCKS
Type_in: CHYMOTRYPSIN in the separate BLOCKS browser search area!
Click: on BLOCKS browser Edit menu option and then type in
TRY1 at FIND option!
Observe: following sequence profile: ( 43) GGSLINDQWVVSAAHC
Click: on boxes bellow GGSLINDQWVVTAAHC sequence, starting at
position B#43.
|
Mouse Mark: residues GGSLINDQWVVTAAHC by sliding mouse above
these residues in the (LF) with left mouse button
pressed.
|
Observe: Identify visually 3D position of this stretch! Obviously,
one can see that very preserved CONTIGUOUS stretch of the sequence
within CHYMOTRYPSIN family of proteins, is not participating
in Interface Forming (IF) area! Only initial and ending residues are
3D_positioned within IF moiety. Thus, STING helped us to quickly
locate BLOCK of the conserved sequence stretch out of IF area!
Final Screen: You will get screen as shown below:
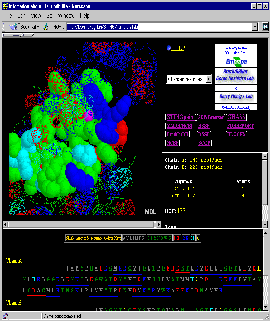
- And Beyond: Use [Interface: 1st half] and
[Interface: 2nd half] to get exclusive view on first half and then
second half of the complementary Interface Forming Residues (IFR)!
Continuing, use [Interface: 1st half] to get
again exclusive look at the first half of the interface...
Such iterative procedure allows users to get better glimpse at IFR.
Very useful STING command will be [Charged residues]: user can see
how charge is distributed and how complemented at Interface halves!
Use also[HOH + 1st half] and [HOH + 2nd half] to get
again exclusive view on first half and then second half of the complementary
Interface Forming Residues (IFR), but now with water molecules in their
vicinity! See FEATURES chapter for
explanation of this command, as well as differences with [Interface: 1st half] and
[Interface: 2nd half]!
|
 STING: Charge complementarity on molecular interfaces
STING: Charge complementarity on molecular interfaces
 STING: Protein/DNA Interface
and Structural Waters
STING: Protein/DNA Interface
and Structural Waters
 STING: Ligand Pocket and Coordinating Residues
STING: Ligand Pocket and Coordinating Residues
 STING: Enzyme/Inhibitor Interface and
Structural Waters
STING: Enzyme/Inhibitor Interface and
Structural Waters